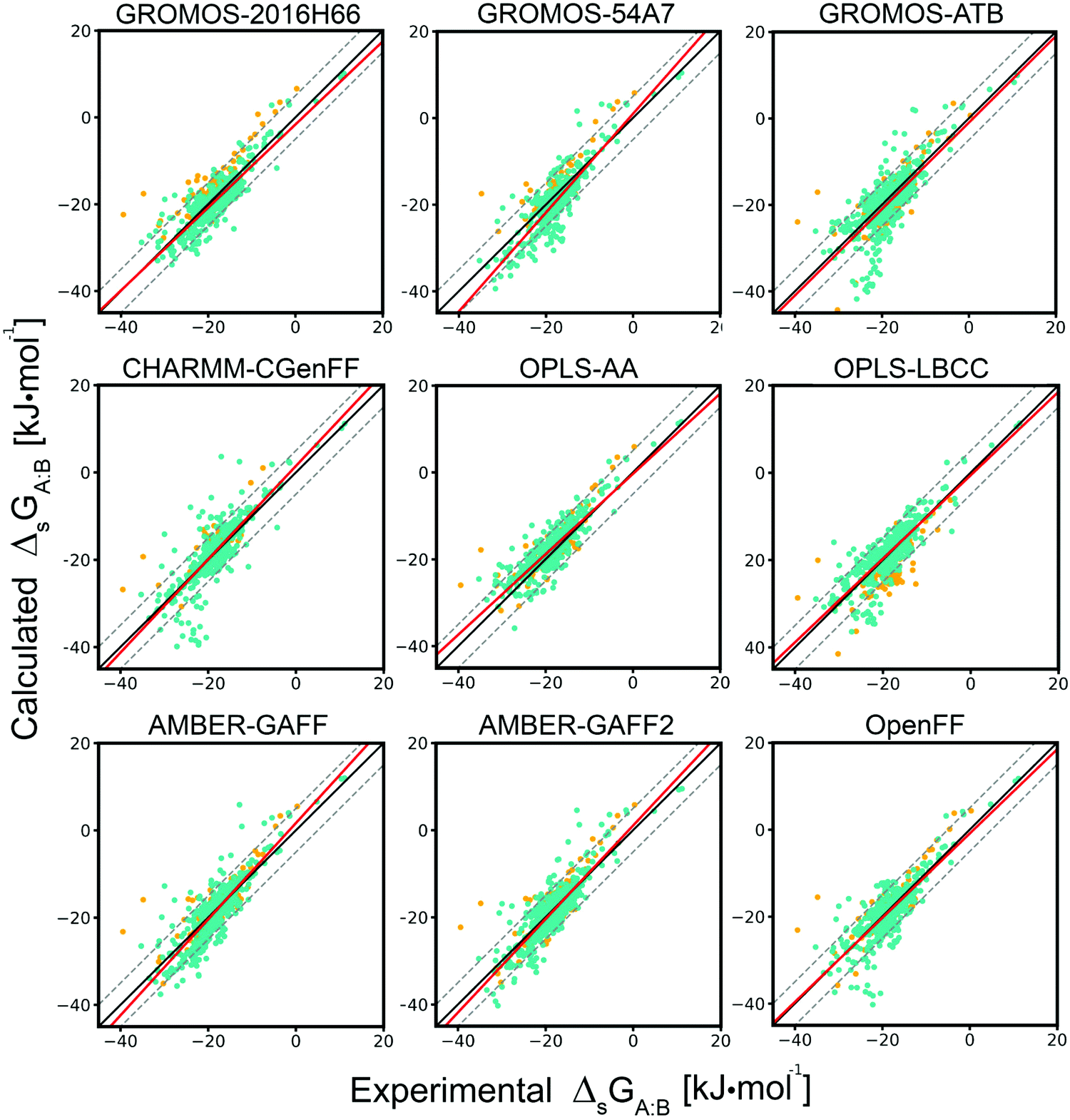
Evaluation of nine condensed-phase force fields of the GROMOS, CHARMM, OPLS, AMBER, and OpenFF families against experimental cross-solvation free ener ... - Physical Chemistry Chemical Physics (RSC Publishing) DOI:10.1039/D1CP00215E

Validation of the Generalized Force Fields GAFF, CGenFF, OPLS-AA, and PRODRGFF by Testing Against Experimental Osmotic Coefficient Data for Small Drug-Like Molecules | Journal of Chemical Information and Modeling

Full article: Free energy perturbation calculations of tetrahydroquinolines complexed to the first bromodomain of BRD4
Assessing the quality of absolute hydration free energies among CHARMMcompatible ligand parameterization schemes

Protein-ligand complex | Gromacs tutorial | CHARMM forcefield | Molecular dynamics simulation - YouTube

Calculations of Absolute Solvation Free Energies with Transformato─Application to the FreeSolv Database Using the CGenFF Force Field | Journal of Chemical Theory and Computation

Protein-ligand complex | Gromacs tutorial | CHARMM forcefield | Molecular dynamics simulation - YouTube
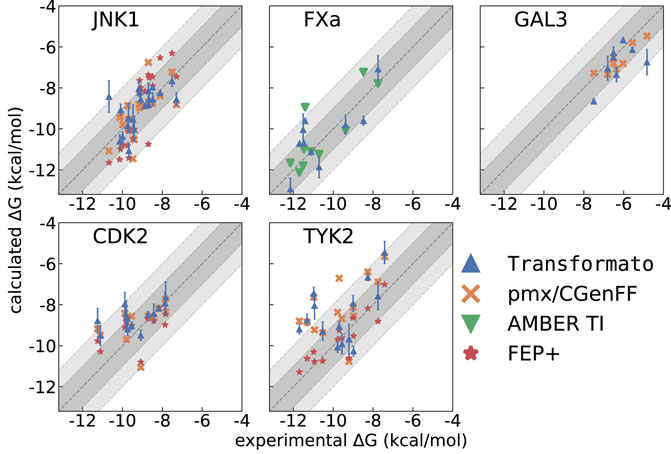
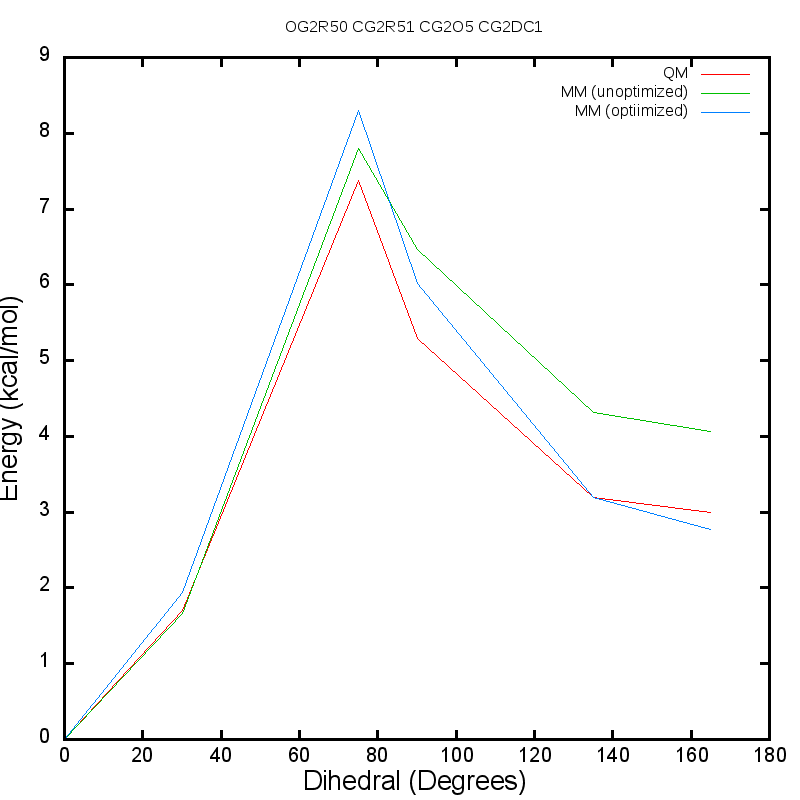
Frontiers | Relative binding free energy calculations with transformato: A molecular dynamics engine-independent tool
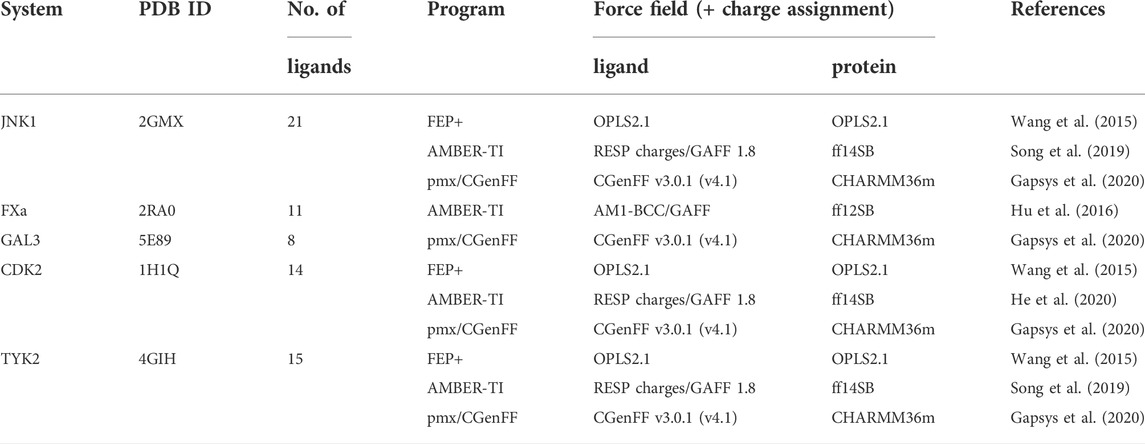
Frontiers | Relative binding free energy calculations with transformato: A molecular dynamics engine-independent tool
molecular dynamics - Alternative to CGenFF for generating large ligand topology - Matter Modeling Stack Exchange

Problem in file (Broken structure ini.pdb) generated using CGenFF for proceeding in MD Simulation - User discussions - GROMACS forums
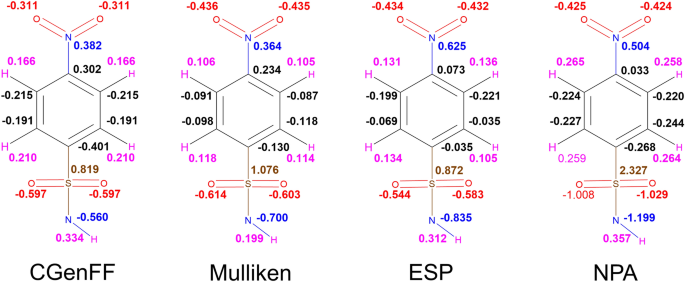
MM/GBSA prediction of relative binding affinities of carbonic anhydrase inhibitors: effect of atomic charges and comparison with Autodock4Zn | Journal of Computer-Aided Molecular Design
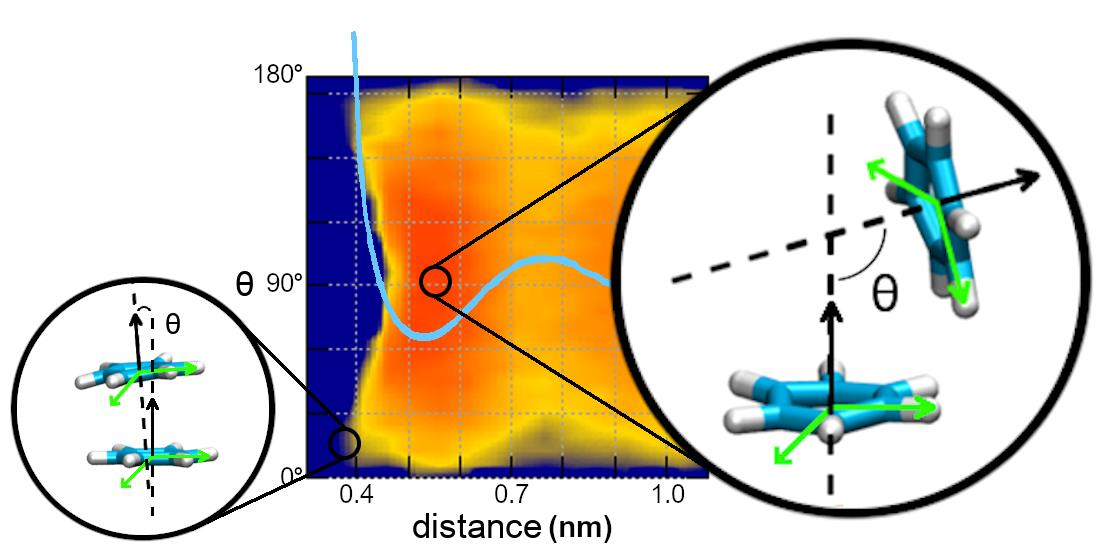
Molecules | Free Full-Text | Comparing Dimerization Free Energies and Binding Modes of Small Aromatic Molecules with Different Force Fields

FFParam: Standalone package for CHARMM additive and Drude polarizable force field parametrization of small molecules - Kumar - 2020 - Journal of Computational Chemistry - Wiley Online Library